The NOE (Nuclear Overhauser Effect) experiment allows for the estimation of through space proton-proton distances. The two dimensional version will capture all possible pairs of protons, but can be lengthy and lack resolution.
A one dimensional version of the experiment allows to obtain a set of distances to a selected proton in shorter time and full resolution of a normal proton spectrum. However, setup of a spectrum requires knowledge of desired shifts from a previous proton spectrum.
On the Avance-500, the parameter set noe1d can be used for 1D NOE.
AUTOMATION
The setup assumes that you already took a 1D proton spectrum and know the ppm values of the signals you want to irradiate. For a procedure where you interactively select several signals from an existing spectrum see the manual setup below.
- Select noe1d as experiment from the pull down menu
- In the parameter section (=) set cnst21 to the ppm value of the signal you want to irradiate.
- Change o1p and sw if required. IF the sample is dilute, you may need to increase ns. For macromolecules (polypeptides, polymers) you may need to decrease the mixing time d8 to a smaller number (0.2s).
- Repeat for as many signals you want to irradiate
In the final spectrum, the irradiated signal is the largest peak. apk usually phases that peak positive, but one needs to think of that peak as an inverted (negative) signal. Hence negative NOEs will show as positive signals, and positive NOEs as negative signals.
MANUAL Setup
- Using the noe1d parameter file
In manual setup, the parameter set noe1d can be used the same way as in automation by entering the ppm value of the signal to be irradiated.
- Using the Topspin builtin interactive setup.
The Topspin builtin interactive setup may be more user friendly, in particular if you want to irradiate multiple signals.
In the [Acquire] flow bar, select [Setup 1D Epts.] from the [More] pulldown menu at the right:

A new flowbar will appear. Select “Define Regions” first.
![]()
This will enter the integrations mode. You now need to define integral regions for each signal you want to irradiate. The width of the integral will correspond to the selectivity of the irradiation.
If you had integrals in the spectrum before (manually of from abs), you need to delete them first. First, select all integrals![]()
Then, click the “Delete Selected” button
![]()
Now integrate only the signals to be irradiated as described in the basic NMR manual.
When done, select Save As and select Save Regions to ‘reg’ option

Now you are ready to click on [Create Datasets] in the 1D Selective Experiment Setup flowchart.

Select “Selective Gradient NOESY” from the pulldown menu. For ROESY or TOCSY, select the respective gradient selective experiment.
You will be prompted to change any parameters if required. Continue with [Accept]
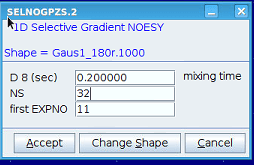
Start the experiment(s) with [OK]. Selecting [Cancel] will create the datasets for you to run manually at a later time.

Click the <-Back button to exit the selective Experiment Toolbar.
![]()