https://uwm.edu/freshwater/mclellan-labs-covid-19-tracking-in-the-news/
Author Archives: dila
Looking ahead on COVID-19

Updates on coronavirus, including an interview with Sandra McLellan on monitoring coronavirus outbreaks through city sewage:
• Our grad students featured at Freshwater Sciences
• Our former postdoc Jen Fisher and her student research assistants
• Stayin’ alive: Long-surviving E. coli strains
Read Ph.D. student Natalie Rumball’s interview in the new issue of the Aquatic Sciences Chronicle: Stayin’ alive: Long-surviving E. coli strains make identifying contaminated beaches more complex than previously thought
• Natalie Rumball chats about E. coli on NPR Lake Effect
 Scroll down to August 3, 2020 to listen to Natalie’s interview!
Scroll down to August 3, 2020 to listen to Natalie’s interview!• Statewide COVID-19 Surveillance
Fox 11 interview on work we are doing with NEW Water in Green Bay, WI

• NPR interview with Sandra McLellan – Surveillance of COVID-19 in sewage

Ryan Newton and Sandra McLellan Photo credit: Troye Fox/UWM Photo Service
Susan Bence interviews Sandra McLellan for WUWM 89.7 Milwaukee NPR – “Because we’ve done sewage surveillance for bacteria for so long, when we started hearing reports that it might be in sewage, that was the first thing that came to mind — of course, we should be sampling sewage,” McLellan says.
The lab is working towards setting up a state wide effort to look for signs of COVID-19 in wastewater samples from treatment plants around Wisconsin. Read or listen to the interview.
• A successful thesis defense for Shuchen Feng, Ph.D.
Development of Indicators for Human Fecal Pollution Using Deep-Sequencing of Microbial Communities
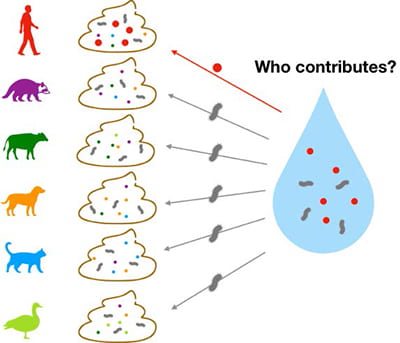
Shuchen Feng’s work was focused on expanding the view of human-associated fecal organisms using deep sequencing of sewage and animal fecal microbial communities. Dr. Feng developed highly specific and sensitive qPCR assays to increase the accuracy of microbial source tracking of human fecal contamination.
Human fecal pollution is a persistent issue of microbial water quality in the U.S. and worldwide. Over the past two decades, microbial source tracking (MST) methods have been developed to identify the presence of human-associated fecal microorganisms. The standard approach is to use quantitative polymerase chain reaction (qPCR) assay to amplify distinct sequences (i.e., markers) of these microorganisms that serve as fecal indicators. To date, most bacterial human fecal marker assays are limited to the genus Bacteroides, leaving a large fraction of fecal microorganisms unexplored. Furthermore, specificities and sensitivities of these assays are negatively affected by animal cross-reactions and varying Bacteroides abundances in the host sources, respectively. My work has been focused on expanding the view of human-associated fecal organisms using deep sequencing of sewage and animal fecal microbial communities. Multiple human-associated alternative indicators were selected from family Lachnospiraceae. Indicators within the genus Bacteroides that were uniquely present in urban sewer infrastructure, but not human or animal gut microbiomes were also identified. Subsequently, qPCR assays were developed for their practical applications to track human fecal source. In addition, mechanisms for low-level animal cross-reactions were explored, further demonstrating host specificities of these new indicators.
• Dr. Jonathan Patz from Global Health Institute, UW-Madison visited the McLellan Lab
Dr. Jonathan Patz from Global Health Institute, Wisconsin-Madison visited the McLellan lab to discuss climate, water and health, and toured the UW Milwaukee – School of Freshwater Sciences.Dr. Patz visited the estuary site where the Mclellan lab has sampling equipment deployed to quantify the amount of untreated sewage that enters the lake for failing infrastructure such as leaking pipes or illicit connections. Dr. Patz visited the estuary site where the Mclellan lab has sampling equipment deployed to quantify the amount of untreated sewage that enters the lake for failing infrastructure such as leaking pipes or illicit connections.
Dr. Patz visited the estuary site where the Mclellan lab has sampling equipment deployed to quantify the amount of untreated sewage that enters the lake for failing infrastructure such as leaking pipes or illicit connections.

Dr. Patz and Dr. McLellan recall their sampling adventures from a few years ago. Data from the Mclellan lab is shown in the article Climate change and waterborne disease risk in the Great Lakes region of the U.S. (Am J Prev Med. 2008 Nov;35(5):451-8), authored by Patz JA*, Vavrus SJ, Uejio CK, McLellan SL.
Ryan Bartelme and Dr. Patz discuss alternative protein sources and recirculating aquaculture systems that move us towards sustainable food production.
