Our overall goal is to change the way we monitor for waterborne pathogens. This framework is outlined in a review article Trends in Microbiol 22:697-706
Microbial Community Profiles Identify New Indicators of Waterborne Pathogens.
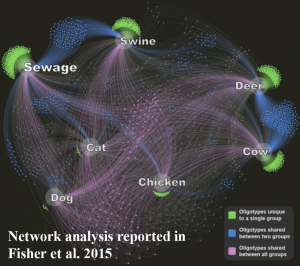 Fecal pollution and its associated pathogens are ubiquitous in urban waters. These waterborne pathogens are estimated to cause between 12 and 16 million cases of gastroenteritis in the US each year. Currently, the growth of fecal indicator organisms such as E. coli or enterococci is used to monitor fecal pollution in recreational environments. However, these indicators are not specific to humans and are abundant in all warm-blooded animals, therefore they are not a good predictor of human pathogen occurrence or an indicator of pollution source. The ability to identify pollution sources is particularly important in a time of limited economic resources, as it carries the potential to create tools that will more effectively direct resources to the most serious sources of fecal pollution. We are taking an entirely new approach for pathogen surveillance by using molecular DNA methods including new sequencing technologies to characterize the complex microbial signatures of fecal pollution and focusing on new fecal indicator groups specific to humans.
Fecal pollution and its associated pathogens are ubiquitous in urban waters. These waterborne pathogens are estimated to cause between 12 and 16 million cases of gastroenteritis in the US each year. Currently, the growth of fecal indicator organisms such as E. coli or enterococci is used to monitor fecal pollution in recreational environments. However, these indicators are not specific to humans and are abundant in all warm-blooded animals, therefore they are not a good predictor of human pathogen occurrence or an indicator of pollution source. The ability to identify pollution sources is particularly important in a time of limited economic resources, as it carries the potential to create tools that will more effectively direct resources to the most serious sources of fecal pollution. We are taking an entirely new approach for pathogen surveillance by using molecular DNA methods including new sequencing technologies to characterize the complex microbial signatures of fecal pollution and focusing on new fecal indicator groups specific to humans.
Human-associated Lachnospiraceae
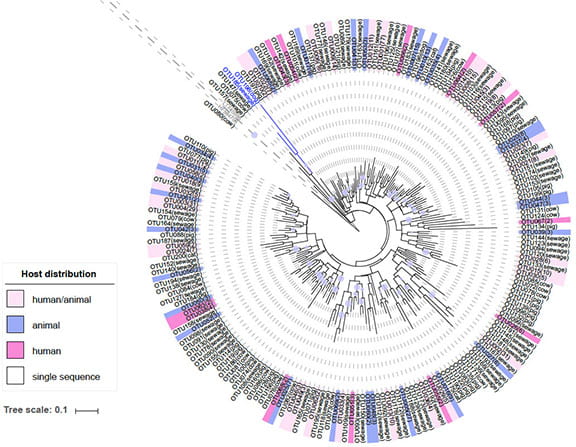 The bacterial family Lachnospiraceae is a promising group for developing human-associated alternate fecal genetic markers. The family Lachnospiraceae comprises up to 15% of the sewage microbial community and has highly diverse 16S rRNA gene sequences among human and different animal hosts. In particular, oligotypes of the genus Blautia reveal unique population structure among different host groups. By determining the V6 region as the most ideal region for developing host-associated fecal markers, we compared the Lachnospiraceae V6 region sequences from sewage and different animal hosts and developed novel alternate fecal marker assays, Lachno3 and Lachno12. Our new Lachnospiraceae assays are highly sensitive (both 100%) to sewage detection through qPCR validations. The Lachno12 assay has cross-reactions with two non-urban animal sources (pig and cow), while Lachno3 assay is considered as strictly human-specific. Combining these human-associated fecal markers with animal-associated fecal markers will provide the highest resolution and specify for assessing fecal pollution sources in urban waters.
The bacterial family Lachnospiraceae is a promising group for developing human-associated alternate fecal genetic markers. The family Lachnospiraceae comprises up to 15% of the sewage microbial community and has highly diverse 16S rRNA gene sequences among human and different animal hosts. In particular, oligotypes of the genus Blautia reveal unique population structure among different host groups. By determining the V6 region as the most ideal region for developing host-associated fecal markers, we compared the Lachnospiraceae V6 region sequences from sewage and different animal hosts and developed novel alternate fecal marker assays, Lachno3 and Lachno12. Our new Lachnospiraceae assays are highly sensitive (both 100%) to sewage detection through qPCR validations. The Lachno12 assay has cross-reactions with two non-urban animal sources (pig and cow), while Lachno3 assay is considered as strictly human-specific. Combining these human-associated fecal markers with animal-associated fecal markers will provide the highest resolution and specify for assessing fecal pollution sources in urban waters.
Sequenced based pipeline to identify sources of pollution
Tracking the source of fecal pollution is essential to assess, prevent, and remediate sanitation issues. In the project funded under National Institutes of Health (NIH) grant, we are developing a bioinformatic pipeline able to recognize, among complex environmental matrices, fecal bacterial signatures originating from human and animal sources.
The pipeline is based on significant differences among fecal bacterial sources, observed through the 16S rRNA gene sequences. The identification of relevant bacterial signatures among sources allow comparison to predict the host(s) responsible for the fecal contamination in an environmental sample. In the future, the pipeline will be available online, as a step forward in microbial source tracking tools.
