From January 2018 to December 2021 the Milwaukee Metropolitan Sewage Distract (MMSD) fellowship funds were used to increase capacity on several projects, that were largely supported by coinciding grants, allowing us to pursue research questions to their fullest. In total ten students were able to perform projects including, but not limited to: examining members of the microbial community that is suspected to influence sewer corrosion such as Arcobacter, completing the Lake Michigan Plume and Hydrodynamic Modeling study, discovering new indicators of fecal pollution, performing preliminary work and research on SARS-CoV-2 surveillance in sewage, and analyzing chloride pollution on local waterways.
A break down on the students wo were helped, and the projects they performed from this funding:
Elexius (Lexi) Passante. Lexi was a graduate research assistant completing her Master’s degree in May 2022. Her primary research project uses salt-loving bacteria (i.e., halophiles) as alternative indicators to assess chloride impaired waters. In the McLellan lab, she also assisted with the SARS-CoV-2 wastewater surveillance project.
Chloride pollution in the Milwaukee River Basin- A Long-term dataset (2021)
Specific study sites within the Milwaukee River Basin were categorized as rural- or urban-impacted based on previous chloride concentration data. Halophiles were isolated from the sediment of sites in each category. Halophiles are extreme bacteria that require the presence of salt to survive which makes them an attractive candidate for determining chloride impaired areas. Chloride or conductivity readings provide a snapshot in time, but monitoring halophiles provides a long-term understanding of how chloride interacts in that ecosystem throughout the year as each freshwater system is unique. A total of 133 samples were collected between February 2020, and February of 2022.
Widespread and permanent Halomonadaceae populations in the Milwaukee River Basin (2021)
Out of all the isolates collected from the study site direct plating samples, a true halophilic family, Halomonadaceae, was isolated 47.4% of the time across all study sites. Of this 47.4%, it consisted of the genera Halomonas, Halovibrio, and Salinicola. Sample analysis illustrates that Halomonadaceae was isolated more frequently and abundantly at the urban sites, that majority of Halomonadaceae isolates collected from our study sites at the cold temperature favored a lower salt concentrations (2% NaCl), and that warm temperature isolates preferred higher salt concentration of (6% NaCl). A further analysis focusing on the prominent genera that were found at the study sites (Halomonas, Halovibrio, and Salinicola) proved Halomonadaceae are permanent members of the microbial community as a result of salting our freshwater resources for decades upon decades.
All current halophilic isolates were sequenced, and merged to construct a phylogenetic tree. Interestingly, the phylogenetic tree showed that certain clades favored salty conditions under both cold and warm whereas others shifted from a higher preferred salt concentration to zero salt in the cold temperature (Figure 1).
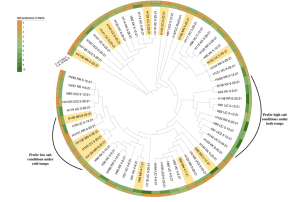
Figure 1. Phylogenetic tree of all current Halomonadaceae isolates collected with their preferred salt concentration (% NaCl (w/v)) at either 18 degrees C (warm) or 5 degrees C (cold). The preferred salt concentration was determined by taking the maximum OD600 absorbance value observed. Phylogenetic tree was constructed in the Interactive Tree of Life (iTOL) software.
Community awareness and outreach on salting our freshwater systems (2021)
Lexi has presented her research at various conferences where she raised awareness of road salt pollution. In October 2021, she was awarded the Cooperative Institute for Great Lakes Research (CIGLR) Engagement, Career Training and Outreach and Communications (ECO) award to increase awareness of chloride’s impact on our environment. With this award, Lexi has filmed a short video series with the UWM student film crew to document the rationale, progress, and impact of her research (Figure 2).
Lexi also worked alongside WI Salt Wise and a local high school teacher in publishing a lesson plan for teachers. This lesson plan is currently in the revision process to be put on the Data Nuggets website (https://datanuggets.org/about-nuggets-2/). Data Nuggets is an online library for teachers to find lessons on any science related topic that are interactive and allow students to work hands on with real environmental data.
In addition to her research, she has been working with James Price, an Assistant Professor at the Center of Water Policy, to propose a framework for improving the economic valuation to help understand potential missing links between chloride concentrations and ecosystem goods and services that are affected.

Figure 2. UWM film student film crew with Lexi on the R/V Neeskay for the CIGLR ECO award educational video series.
Chloride pollution in the Root River – A short-term dataset (2021)
Lexi collaborated with UWM’s Geoscience Department, Assistant Professor Charles Paradis, on their Root River road salt transport study that is funded by the Wisconsin Sea Grant. In this collaboration, Leah Dechant (graduate student), took sediment samples for Lexi twice a week starting in July 2021 – September 2021 from an urban Root River and rural Root River site near Racine, WI. The Racine sites were screened for the same halophiles, and parallel the results discovered in Lexi’s research.
Melissa Schussman (Master Student). Melissa was a civil and environmental engineering Master’s degree student, working primarily on detecting of SARS-CoV-2 in wastewater. Her works helped local Wisconsin communities grasp the severity of COVID-19, added to the growing body of knowledge surrounding Wastewater Surveillance.
SARS-CoV-2 Wastewater Epidemiology (2021)
Since Septmber 2020, the McLellan labs has been tracking the incidence of COVID-19 from wastewater influent in 8 wastewater treatment plants (WWTPs) across Wisconsin. Data form this project can be found at on the Wisconsin Department of Health Services (DHS) dashboard (Figure 3). Melissa Schussman has taken over from Shuchen Feng and Adelaide Roguet as the lead researcher and coordinator for this project. In 2020-2021, she began weekly monitoring of the evolving various variants of concern and influenza from multiple WWTP’s across Wisconsin to expand our surveillance efforts. As of December 2021, we have processed and analyzed over 3,000 samples.
In addition to monitoring, Melissa has performed data analysis to determine WWTP pipe timelines during wet and dry periods, and the impact this sewer travel time has on the viability of SARS-CoV-2 RNA. To combat degradation seen throughout the sewer system, Melissa has been researching ways to increase surveillance target amplification and detection to ensure wastewater surveillance can continue to provide relevant results even when cases decrease. This research included various techniques such as using primary settled solids samples instead of influent samples, analyzing various concentration methods, and implementing new extraction methods.
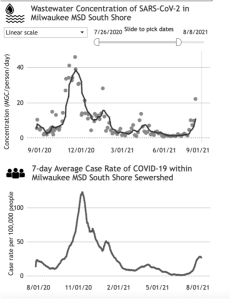
Figure 3. Comparison of wastewater surveillance (top image) versus reported clinical data (bottom image) of COVD-19 in the Milwaukee South Shore WWTP district taken from https://www.dhs.wisconsin.gov/covid-19/wastewater.htm#wastewater dashboard.
Lou LaMartina (Ph.D. Student). Lou LaMartina is a Ph.D. student working on several projects associated with wastewater microbial communities. In 2020, her work focused on understanding the temporal dynamics of bacterial communities in the MMSD sewer system and how changes in the community relate to antibiotic resistance gene loads entering the treatment plants over a year. Her ongoing work will provide a more detailed understanding of whether the seasonal changes in sewer pipe microbial communities impact the load of antibiotic resistance entering treatment plants or if these changes predict bulking/foaming events at the plants.
Seasonal trends in a five-year time series of raw wastewater (2019)
In 2019, we investigated the seasonality of wastewater to examine if microbial communities in influent to wastewater treatment plants were repeated and predictable. Lou LaMartina sequenced microbial community DNA in monthly influent samples from Jones Island and South Shore, spanning a total of five years. Lou discovered that microbial community composition in influent to both Jones Island and South Shore followed seasonal patterns, and that microbes with the strongest seasonal fluctuations were not associated with human waste, but rather may have been persistent residents to pipe systems. In each of the five years analyzed, microbial communities appeared to assemble in two steady states, during spring and fall (Figure 4). Our labs are also interested in impacts wastewater can have on human health, directly or indirectly. Wastewater is believed to be a hotspot for the evolution of bacteria that are resistant to antibiotics. By targeting genes responsible for antibiotic resistance and track them in the environment, Lou discovered from this same sample set that antibiotic resistance genes appear to have seasonal trends that are coupled with microbial communities in the fall steady state. Meaning, antibiotic resistance gene concentrations are higher in fall than any other time of year.
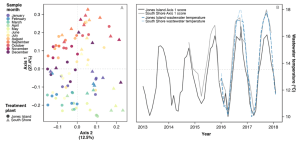
Figure 4. (A) Principal coordinate analysis (PCoA) of influent bacterial communities from two Milwaukee WWTPs sampled once a month for five years. (B) PCoA Axis 1 scores from both WWTPs over time (solid grey lines) plotted with wastewater temperatures (blue dashed lines).
Characterize sewer pipe bacteria across geographic locations (2020)
In 2020 Ms. LaMartina used samples from the above analysis in addition to ones from other states across the US to determine that Milwaukee bacterial communities were very similar to the communities from other northern US city sewer systems, but were distinct from systems in southern US cities, again pointing to temperature as a driver of what bacteria are found in sewer systems. Overall we now have a good method for predicting the bacterial community composition entering municipal wastewater treatment plants. In the coming months, Ms. LaMartina will be working with Dr. McNamara’s group at Marquette to identify if there is any relationship between changes in the treatment plant influent communities and those in the activated sludge tanks.
Adelaide Roguet (Postdoctoral Researcher). Dr. Roguet earned her Ph.D. at University of Paris-Est, specializing in public health and microbial ecology. Currently she develops bioinformatics resources and investigates diversity and fate of abundant bacterial genera in untreated sewage.
Pipeline for microbial source tracking (2018-2019)
Adelaide Roguet developed a microbial bioinformatics pipeline that works as a faster source tracking method to perform health risk assessment. This pipeline that was trained and validated from Milwaukee’s beaches and rivers, integrates a machine learning approach used to accurately identify human/animal sources responsible for fecal pollution in recreational waters (Figure 5).
With collaborators from the University of Chicago, a web interface to help scientists and managers define the source of bacterial pollution as terrestrial runoff or sediment resuspension in hydrologic plumes, to help assess, prevent, and remediate sanitation issues was implemented by Adelaide Roguet. This online platform named FORENSIC (FORest Enteric Source IdentifiCation, http://forensic.sfs.uwm.edu/), makes water quality assessments accessible to public health and water resources organizations, thus help to assess, prevent, and remediate sanitation issues. In the Milwaukee River watershed, FORENSIC will help to quickly screen the potential source(s) responsible for bacterial pollution in terrestrial runoff or sediment resuspension.

Figure 5. Flow chart of bioinformatics pipeline. From the left to the middle presents how the pipeline is ‘trained’. From the right to the middle presents how a given environmental sample is identified as contaminated by a specific source.
Arcobacter, a predominant bacterial group in sewage (2019)
Arcobacter has been identified worldwide as one of the most abundant bacterial genera in untreated sewage. Arcobacterholds about 30 species, among which, Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii, are considered as potential human pathogens. In the past few years, our lab isolated hundreds of isolates from Jones Island and South Shore influent (Figure 6). And, to allow comparison of species genome content from two distinct geographic locations we isolated Arcobacter strains from Hawaii in late 2018 to early 2019. A total of 13 genomes from Milwaukee and Honolulu have been sequenced and assembled. Adelaide started to investigate the ecological reasons of the co-occurrence of Arcobacter and Acinetobacter (the second most abundant genus in sewage), and to what extent their populations are temperature-driven. In the future, we plan to conduct a finer analysis to better understand their ecology and the reasons they thrive and propagate in sewage is in progress.

Figure 6. Arcobacter isolation from sewage using blood agar plates.
Shuchen Feng (Ph.D. Student and Postdoctoral Researcher). Shuchen Feng completed her Ph.D. in the McLellan lab during the summer of 2019, and continued to work as a postdoctoral researcher in the lab. Dr. Feng is specializing in development and validation of high-specificity genetic markers used for microbial source tracking. Additionally, she started working in SARS-CoV-2 sewage surveillance when the COVID-19 epidemic began in March 2020.
Sewage specific Bacteroides markers development (2018)
Our lab has been working on identifying fecal markers and assays to trace human fecal pollution in water environments. In the past year of 2018, Shuchen Feng developed two new marker assays (BacV4V5-1 and BacV6-21 assays) from one of the most abundant groups of human fecal bacteria, genus Bacteroides. These two marker assays were proved to target a sewage derived Bacteroides organism, which very likely originates from a sewer pipe, rather than the normally considered human fecal source. Additionally, the BacV4V5-1 and BacV6-21 marker assays could add another layer of evidence for sewage loading quantification in water environment with low or no impact from possible animal fecal sources.
Sewage specific bacterial markers (2019)
In 2019, Shuchen Feng identified ecological and technical mechanisms for cross-reaction of human/sewage genetic markers with non-targeted animal hosts. For this work, a total number of 365 raw fecal samples from 22 types of animals were collected from the United States and Australia. By analyzing more than 130 million DNA sequences from these samples, Shuchen found that some true cross-reactions of human/sewage genetic markers were associated with atypical community compositions of organisms within the bacterial genera Blautia and Bacteroides. The atypical communities could be associated with diet and cohabitation of animal hosts (Figure 7).
Also, Shuchen compared the frequency of qPCR cross-reaction to the frequency of marker identification in DNA sequencing. She found that the lower specificity in qPCR assays (compared to sequence data) were mainly caused by qPCR amplification of sequences that were highly similar to the marker genes, but were not occurrences of the exact marker genes in animal feces. This work demonstrated that a combination of multiple human/sewage marker assays from different microorganisms should be used to increase the confidence of human fecal source detection, which is critical to diagnose and remediate sanitation concerns caused by human fecal pollution in water environment.

Figure 7. Non-metric multidimensional scaling (NMDS) analysis of microbial communities of all mammal samples sequenced in this study (n =219).
Investigate propagation, adaptation and influence of Escherichia coli in sewage (2020)
Sewers are hotspots for antibiotic resistance genes and antibiotic resistant pathogens, but the overall contribution of this reservoir to the dissemination of antibiotic resistance to humans is unknown. In 2020, Shuchen Feng did research exploring the fate of antibiotic resistant E. coli in the sewer environment.
Shuchen constructed an environmental E. Coli strain library that consists of > 1,000 E. Coli strains from freshly sampled residential sewage, sewer pipe sediments and biofilms. Genetic phylogroups of all the E. coli strains, and antibiotic resistance profiles using 7 antibiotics, including clinically important carbapenems, for all the strains were identified from this sample set. A total of 471 strains were identified that are resistant to one or more antibiotics, with 30 strains that are carbapenem resistant and showed resistance patterns to almost all the tested antibiotics. For more detailed sub-typing of antibiotic resistant E. Coli strains, Shuchen Feng simultaneously sequenced important E. coli house-keeping genes, and E. coli taxonomic identification DNA to obtain more definitive sequencing results.
SARS-CoV-2 Wastewater Surveillance (2020-2021)
Since mid-March 2020, Dr. Feng has been working on the SARS-CoV-2 wastewater surveillance project in Wisconsin. Her early efforts in SARS-CoV-2 surveillance included developing and optimizing methods for virus RNA concentration, extraction and digital PCR amplification (Figure 8). These early optimization efforts set the technical basis for our on-going real-time SARS-CoV-2 wastewater surveillance project.

Figure 8. Shuchen Feng setting up PCR reaction to analyze SARS-CoV-2 concentration from sewage.
Jill McClary (Postdoctoral Researcher): Dr. McClary is a civil and environmental engineer, earning her Ph.D. at Stanford University. She specializes in the detection, fate, and public health impact of microbial pollutants in natural and engineered water systems.
Plume Study (2018-2020)
Three labs at the School of Freshwater Sciences are collaborating on a project focused on variable aspects of water quality associated with Milwaukee watershed plumes delivered to Lake Michigan. Jill McClary, a recent postdoc in the Newton and McLellan labs, worked alongside the Klump lab to collected approximately 100 sediment samples (Figure 9) and approximately 300 water samples at various locations along the Milwaukee River and within the Milwaukee Harbor, help determine the sources and timing of rain-driven pollution events in Milwaukee River plumes.

Figure 9. Collection of sediment from traps deployed in the Milwaukee River near Saukville.
In 2019, a subset of approximately 100 samples, and approximately 50 sediment samples collected during base-flow, rain, and CSO events were analyzed for an in-depth microbial and sediment characterization to determine the sources of both microbial and sediment pollutants in order to recommend appropriately targeted strategies for their mitigation. This analysis suggest equal contributions of urban and rural sources to sediment pollution in the Milwaukee Harbor Estuary.
And in 2020, Jill McClary organized the completion of this project, including a project extension to incorporate additional hydrodynamic modeling efforts. She worked with Bahram Khazaei (PhD student / modeling engineer), Dr. Hector Bravo (engineering professor), and Emily Koster (MS student) to generate model simulations of FIB, TSS, and human fecal marker fate in nearshore Lake Michigan for six different high flow events (Figure 10). She also completed DNA sequencing of 200 samples and conducted analysis to identify DNA sequences associated with sediment that may be used in TSS source tracking.

Figure 10. Hydrodynamic model snapshots at a Milwaukee River sampling site of fecal coliform, E. coli, and TSS impacts on the Lake Michigan nearshore from June 2018 rain + CSO event.
Caleb Abfall (Undergraduate Research Assistant)
PECO disinfection set up from Solar Water Works (2018)
The McLellan Lab has an ongoing collaboration with Solar Water Works, a company based out of the Global Water Center in Milwaukee. Caleb Abfall was the primary student collecting and processing samples from three newly installed SWW disinfection set ups. Caleb collected water samples and ran microbial plate count assays to analyze the effectiveness of the solar powered PECO equipment at removing contaminating fecal bacteria (E. coli, enterococci and fecal coliforms) from stormwater runoff. Contaminated runoff is disinfected as it flows through filtering panels seen in Figure 11. The preliminary field work that Caleb completed in 2018 shows PECO filtration significantly reduced the number of fecal bacteria in filtered samples. At the Bradford Beach site, filtered samples had E. coli counts that were 10 to 100-fold reduced from source counts.

Figure 11. Stormwater recycling fountain with PECO solar filtration panels set up at Freshwater Plaza Apartments.
South Shore monitoring (2018)
The South Shore Beach improvement project is poised to move the swimming beach, which is often closed due to high counts of fecal bacteria, to a new location south of where it currently sits. The McLellan lab has found the new beach location to have consistently cleaner water. Caleb Abfall has been actively involved with water sampling at the current South Shore beach and at the proposed beach. Additionally, a new green infrastructure parking lot drainage system has been installed and functioning over the 2018 summer swim season (Figure 12). Lower stormwater runoff volume from the new parking lot may have reduced bacterial beach contamination.

Figure 12. Parking lot improvements at South Shore Beach that include a new parking lot bioswale and drain and rain garden near Lake Michigan.
